| # Install on Terminal of MacOS # 1. pandas #pip3 install -U pandas # 2. NumPy #pip3 install -U numpy # 3. matplotlib #pip3 install -U matplotlib # 4. scikit-learn (sklearn) #pip3 install -U scikit-learn # 5. statsmodels #pip3 install -U statsmodels |
1_MacOS_Terminal.txt
| ########## Run Terminal on MacOS and execute ### TO UPDATE cd "YOUR_WORKING_DIRECTORY" python3 stpkg00.py #python3 stpkg01.py python3 stpkg01.py yX.csv y python3 stpkg02.py yXSTD.csv #Multiple Linear Regression python3 stpkg03.py yXSTD.csv y x1 x2 x3 #Polynomial SINGLE Linear Regression python3 stpkg04.py yXSTD.csv y x1 3 |
Input data files
N/A
| NaN |
Python files
stpkg00.py
| #################### Data Generation #################### ''' x1raw = σ(1,1) x2raw = σ(2,2) x3raw = x1 + σ(2,2) x1 = σ(0,1) x2 = σ(0,1) x3 = σ(0,1) y = 1.0 + (2.0 * x1) + (0.5 * x2) + (0.1 * x3) + (0.05 * x1 * x2) + (0.2 * (x1)^2) + (0.02 * (x1)^3) + σ(0,1) ''' import scipy as sp import pandas as pd from sklearn.preprocessing import StandardScaler #import math import numpy as np sp.random.seed(0) #numbe of samples n = 100 x1 = sp.random.normal(1,1,n) x2 = sp.random.normal(2,2,n) x3 = x1 + sp.random.normal(2,2,n) # #print(x1) #[ 2.76405235 ... #print(type(x1)) #<class 'numpy.ndarray'> # #print(x2) #[ 5.76630139e+00 ... # #print(x3) #[ 4.02568867 ... x1 = pd.DataFrame(x1) x1 = x1.rename(columns={0: 'x1'}) #print(x1) x2 = pd.DataFrame(x2) x2 = x2.rename(columns={0: 'x2'}) #print(x2) x3 = pd.DataFrame(x3) x3 = x3.rename(columns={0: 'x3'}) #print(x3) X = pd.concat([x1, x2, x3], axis=1) #print(X) #print(X.describe()) #standard deviation (sample, NOT population) X.to_csv('X.csv', header=True, index=False) #scaler = StandardScaler() scaler = StandardScaler() scaler.fit(X) #print(scaler.mean_) # mean #[1.05980802 2.16402594 2.94134349] # #print(scaler.var_) # variance #print(type(scaler.var_)) #<class 'numpy.ndarray'> #print((scaler.var_) ** 0.5) # standard deviation (population) #[1.00788224 2.06933399 2.22108027] XSTD = pd.DataFrame(scaler.transform(X)) XSTD = XSTD.rename(columns={0: 'x1', 1: 'x2', 2: 'x3'}) XSTD.to_csv('XSTD.csv', header=True, index=False) #print(XSTD) ''' x1 x2 x3 0 1.690916 1.740790 0.488206 1 0.337687 -1.381867 -0.008980 2 0.911743 -1.307182 1.457270 3 2.164028 0.857652 1.625370 4 1.793612 -1.213082 1.443657 .. ... ... ... 95 0.641707 -0.245064 1.368259 96 -0.048922 0.666666 0.119134 97 1.712564 0.716647 1.355392 98 0.066579 2.011491 -0.276140 99 0.339505 1.212482 0.540619 [100 rows x 3 columns] # print(XSTD.describe()) ''' ''' x1 x2 x3 count 1.000000e+02 1.000000e+02 1.000000e+02 mean -1.143530e-16 1.576517e-16 -5.440093e-17 std 1.005038e+00 1.005038e+00 1.005038e+00 min -2.592364e+00 -2.228172e+00 -2.412135e+00 25% -6.981616e-01 -7.997189e-01 -6.266296e-01 50% 3.401995e-02 -5.543630e-02 1.232380e-02 75% 6.719727e-01 7.398196e-01 6.364589e-01 max 2.192663e+00 2.224031e+00 2.300629e+00 ''' #print(XSTD['x1']) #print(type(XSTD['x1'])) #<class 'pandas.core.series.Series'> # #print(np.array(XSTD['x1'])) #print(type(np.array(XSTD['x1']))) #<class 'numpy.ndarray'> tmpy = (2.0 * np.array(XSTD['x1'])) tmpy = tmpy + (0.5 * np.array(XSTD['x2'])) tmpy = tmpy + (0.1 * np.array(XSTD['x3'])) tmpy = tmpy + (0.05 * np.array(XSTD['x1']) * np.array(XSTD['x2'])) tmpy = tmpy + (0.2 * (np.array(XSTD['x1']) ** 2)) tmpy = tmpy + (0.02 * (np.array(XSTD['x1']) ** 3)) tmpy = tmpy + sp.random.normal(0,1,n) tmpy = tmpy + 1.0 #print(tmpy) #print(type(tmpy)) #<class 'numpy.ndarray'> y = pd.DataFrame(tmpy) y = y.rename(columns={0: 'y'}) y.to_csv('y.csv', header=True, index=False) #print(y) #print(type(y)) #<class 'pandas.core.frame.DataFrame'> #y = 1.0 + (2.0 * x1) + (0.5 * x2) + (0.1 * x3) + (0.05 * x1 * x2) + (0.2 * (x1)^2) + (0.02 * (x1)^3) + σ(0,1) #y = 1.0 + (2.0 * x1) + (0.5 * x2) + (0.1 * x3) + (0.05 * x1 * x2) + (0.2 * (x1**2)) + (0.02 * (x1**3)) + sp.random.normal(0,1,n) #y = 1.0 + (2.0 * 1.690916) + (0.5 * 1.740790) + (0.1 * 0.488206) + (0.05 * 1.690916 * 1.740790) + (0.2 * (1.690916**2)) + (0.02 * (1.690916**3)) + 0 #print(1.0 + (2.0 * 1.690916) + (0.5 * 1.740790) + (0.1 * 0.488206) + (0.05 * 1.690916 * 1.740790) + (0.2 * (1.690916**2)) + (0.02 * (1.690916**3)) + 0) #6.11675670334485 yX = pd.concat([y, x1, x2, x3], axis=1) yX.to_csv('yX.csv', header=True, index=False) |
stpkg01.py
| #################### Data Loading and Standardization ########## data file(s) to load # yX.csv (which includes dependent variable y and indepdent variables X: x1, x2, ...) ########## ########## Run this code as follows #python3 stpkg01.py yX.csv y #python3 stpkg01.py (data file to load) (dependent variable/target in yX.csv) ########## import import sys import pandas as pd from sklearn.preprocessing import StandardScaler #print(sys.argv[0]) #stpkg01.py # dfname = sys.argv[1] yname = sys.argv[2] ########## ########## loading data #yX = pd.read_csv('yX.csv', header = 0) yX = pd.read_csv(dfname, header = 0) # print(yX) #print(yX.describe()) #X = yX.drop(['y'], axis=1) X = yX.drop([yname], axis=1) #print(X) X.to_csv('X.csv', header=True, index=False) #y = yX['y'] y = yX[yname] y.to_csv('y.csv', header=True, index=False) scaler = StandardScaler() scaler.fit(X) #print(scaler.mean_) # mean #[1.05980802 2.16402594 2.94134349] # #print((scaler.var_) ** 0.5) # standard deviation (population) #[1.00788224 2.06933399 2.22108027] XSTD = pd.DataFrame(scaler.transform(X), columns = X.columns) print(XSTD.describe()) ''' x1 x2 x3 count 1.000000e+02 1.000000e+02 1.000000e+02 mean -1.187939e-16 1.620926e-16 -2.320366e-16 std 1.005038e+00 1.005038e+00 1.005038e+00 min -2.592364e+00 -2.228172e+00 -2.412135e+00 25% -6.981616e-01 -7.997189e-01 -6.266296e-01 50% 3.401995e-02 -5.543630e-02 1.232380e-02 75% 6.719727e-01 7.398196e-01 6.364589e-01 max 2.192663e+00 2.224031e+00 2.300629e+00 ''' XSTD.to_csv('XSTD.csv', header=True, index=False) yXSTD = pd.concat([y, XSTD], axis=1) yXSTD.to_csv('yXSTD.csv', header=True, index=False) |
stpkg02.py
| #################### Initial Data Analysis ########## Run this code as follows #python3 stpkg02.py yXSTD.csv #python3 stpkg02.py (data file to load) ########## import import sys import pandas as pd from pandas.plotting import scatter_matrix import matplotlib.pyplot as plt import seaborn as sns #print(sys.argv[0]) #stpkg02.py # dfname = sys.argv[1] ########## ########## loading data #yXSTD = pd.read_csv('yXSTD.csv', header = 0) yXSTD = pd.read_csv(dfname, header = 0) #print(yXSTD.describe()) ''' y x1 x2 x3 count 100.000000 1.000000e+02 1.000000e+02 1.000000e+02 mean 1.008893 -1.232348e-16 1.576517e-16 -2.364775e-16 std 2.384226 1.005038e+00 1.005038e+00 1.005038e+00 min -4.001685 -2.592364e+00 -2.228172e+00 -2.412135e+00 25% -0.432215 -6.981616e-01 -7.997189e-01 -6.266296e-01 50% 0.755357 3.401995e-02 -5.543630e-02 1.232380e-02 75% 2.321260 6.719727e-01 7.398196e-01 6.364589e-01 max 6.808487 2.192663e+00 2.224031e+00 2.300629e+00 ''' ########## scatter matrix #scatter_matrix(yXSTD, figsize=(6.4, 4.8)) #640 x 480 scatter_matrix(yXSTD, figsize=(800/100, 600/100)) plt.suptitle('Scatter Matrix') plt.savefig("Fig_02_1.png") plt.show() plt.close() ########## correlation cor = yXSTD.corr() #print(cor) ax = sns.heatmap( cor, annot=True, fmt='.4f', #f for fixed number of decimal places (4 in this case); g is for variable numbers vmin=-1, vmax=1, center=0, #cmap='Blues', cmap=sns.diverging_palette(20, 220, n=200), square=True ) ax.set_xticklabels( ax.get_xticklabels(), #rotation=45, horizontalalignment='right' ); plt.suptitle('Correlation Heatmap') plt.savefig("Fig_02_2.png") plt.show() plt.close() |
stpkg03.py
| #################### Multiple Linear Regression ########## Run this code as follows #python3 stpkg03.py yXSTD.csv y x1 x2 x3 #python3 stpkg03.py (data file to load) (dependent variable/target in yXSTD.csv) (3 specified independent variables in yXSTD.csv for multiple linear regression) ########## import import sys import pandas as pd from sklearn.model_selection import train_test_split #from sklearn.linear_model import LinearRegression from sklearn import linear_model import statsmodels.api as sm import matplotlib.pyplot as plt #from pandas.plotting import scatter_matrix #import seaborn as sns #print(sys.argv[0]) #stpkg03.py # dfname = sys.argv[1] yname = sys.argv[2] x1name = sys.argv[3] x2name = sys.argv[4] x3name = sys.argv[5] ########## ########## loading data #yXSTD = pd.read_csv('yXSTD.csv', header = 0) yXSTD = pd.read_csv(dfname, header = 0) #print(yXSTD.describe()) ''' y x1 x2 x3 count 100.000000 1.000000e+02 1.000000e+02 1.000000e+02 mean 1.008893 -1.232348e-16 1.576517e-16 -2.364775e-16 std 2.384226 1.005038e+00 1.005038e+00 1.005038e+00 min -4.001685 -2.592364e+00 -2.228172e+00 -2.412135e+00 25% -0.432215 -6.981616e-01 -7.997189e-01 -6.266296e-01 50% 0.755357 3.401995e-02 -5.543630e-02 1.232380e-02 75% 2.321260 6.719727e-01 7.398196e-01 6.364589e-01 max 6.808487 2.192663e+00 2.224031e+00 2.300629e+00 ''' XSTD = yXSTD.drop([yname], axis=1) #print(XSTD) #print(type(XSTD)) #X.to_csv('XSTD.csv', header=True, index=False) y = yXSTD[yname] y = pd.DataFrame(y) #print(y) #print(type(y)) #y.to_csv('y.csv', header=True, index=False) ########## ########## creating training and test data XSTD_train, XSTD_test, y_train, y_test = train_test_split(XSTD, y, test_size=0.2, random_state = 0) #print(XSTD_train.shape) #print(XSTD_test.shape) #print(y_train.shape) #print(y_test.shape) ''' (80, 3) (20, 3) (80, 1) (20, 1) ''' lr = linear_model.LinearRegression().fit(XSTD_train, y_train) print(f"R2 of Training Data: {lr.score(XSTD_train, y_train):.4}") print(f"R2 of Test Data (based on a model by Training Data): {lr.score(XSTD_test, y_test):.4}") ########## Multiple Linear Regression (with Training Data) smXSTD_train = sm.add_constant(XSTD_train) model = sm.OLS(y_train, smXSTD_train) results = model.fit() print(results.summary()) ''' OLS Regression Results ============================================================================== Dep. Variable: y R-squared: 0.839 Model: OLS Adj. R-squared: 0.832 Method: Least Squares F-statistic: 131.5 Date: Sun, 12 Jul 2020 Prob (F-statistic): 5.23e-30 Time: 18:49:50 Log-Likelihood: -111.79 No. Observations: 80 AIC: 231.6 Df Residuals: 76 BIC: 241.1 Df Model: 3 Covariance Type: nonrobust ============================================================================== coef std err t P>|t| [0.025 0.975] ------------------------------------------------------------------------------ const 1.0679 0.113 9.487 0.000 0.844 1.292 x1 1.8865 0.137 13.767 0.000 1.614 2.159 x2 0.5003 0.111 4.490 0.000 0.278 0.722 x3 0.3011 0.135 2.230 0.029 0.032 0.570 ============================================================================== Omnibus: 0.606 Durbin-Watson: 2.329 Prob(Omnibus): 0.739 Jarque-Bera (JB): 0.733 Skew: -0.178 Prob(JB): 0.693 Kurtosis: 2.694 Cond. No. 1.96 ============================================================================== Warnings: [1] Standard Errors assume that the covariance matrix of the errors is correctly specified. ''' with open('results.summary.txt', 'w') as f: print(results.summary(), file=f) #print(results.params) #print(type(results.params)) #<class 'pandas.core.series.Series'> # #print(results.params.shape) #(4,) # #print(len(results.params)) #4 b0 = results.params['const'] b1 = results.params[x1name] b2 = results.params[x2name] b3 = results.params[x3name] #print(b0) #1.0679105398301059 #print(b1) #1.8865026849796247 #print(b2) #0.5003158682861151 #print(b3) #0.3010874982341183 #print('R2: ', results.rsquared) #R2: 0.8385140722177117 #################### traing data integration #print(pd.concat([y_train, XSTD_train], axis=1)) yXypred_train = pd.concat([y_train, XSTD_train, b0 + (b1 * XSTD_train[x1name]) + (b2 * XSTD_train[x2name]) + (b3 * XSTD_train[x3name])], axis=1) yXypred_train = yXypred_train.rename(columns={0: 'ypred'}) yXypred_train = pd.concat([yXypred_train, yXypred_train[yname] - yXypred_train['ypred']], axis=1) yXypred_train = yXypred_train.rename(columns={0: 'residual'}) yXypred_train.to_csv('yXypred_train.csv', header=True, index=False) #print(yXypred_train.columns) #Index(['y', 'x1', 'x2', 'x3', 'ypred'], dtype='object') ########## plot: training data (x1, y) and predicted data (x1, ypred) #print(max(yXypred_train[x1name])) #2.164027787075764 # #print(min(yXypred_train[x1name])) #-4.001684888412238 # #print(max(yXypred_train[yname])) #6.808486625095364 # #print(min(yXypred_train[yname])) #-2.5923641824877004 plt.figure(figsize=(8, 8)) # plt.scatter(yXypred_train[x1name], yXypred_train[yname], color = 'blue', label='Training Data') # plt.scatter(yXypred_train[x1name], yXypred_train['ypred'], color = 'red', label='Predicted Data') # plt.legend(bbox_to_anchor=(1, 0), loc='lower right', borderaxespad=1, fontsize=10) plt.title(x1name + ": Training Data, " + yname + ": Training Data and Predicted Data") plt.xlabel(x1name) plt.ylabel(yname) plt.grid(True) # plt.text(min(yXypred_train[x1name]), max(yXypred_train[yname]) * 1.00, "y = b0 + (b1 * x1) + (b2 * x2) + (b3 * x3)", size = 10, color = "black") plt.text(min(yXypred_train[x1name]), max(yXypred_train[yname]) * 0.90, "b0 = " + str(b0), size = 10, color = "black") plt.text(min(yXypred_train[x1name]), max(yXypred_train[yname]) * 0.80, "b1 = " + str(b1), size = 10, color = "black") plt.text(min(yXypred_train[x1name]), max(yXypred_train[yname]) * 0.70, "b2 = " + str(b2), size = 10, color = "black") plt.text(min(yXypred_train[x1name]), max(yXypred_train[yname]) * 0.60, "b3 = " + str(b3), size = 10, color = "black") plt.text(min(yXypred_train[x1name]), max(yXypred_train[yname]) * 0.50, "R2 = " + str(results.rsquared), size = 10, color = "black") # plt.savefig('Fig_03_1a.png') plt.show() ########## plot: training data (x1) and residual data (y - ypred) plt.figure(figsize=(8, 8)) # plt.scatter(yXypred_train[x1name], yXypred_train['residual'], color = 'green', label='Residual (actual - predicted)') # plt.legend(bbox_to_anchor=(1, 0), loc='lower right', borderaxespad=1, fontsize=10) plt.title(x1name + ": Training Data, " + 'residual' + ": Residual (actual - predicted)") plt.xlabel(x1name) plt.ylabel('Residual') plt.grid(True) # plt.savefig('Fig_03_1b.png') plt.show() #################### test data integration yXypred_test = pd.concat([y_test, XSTD_test, b0 + (b1 * XSTD_test[x1name]) + (b2 * XSTD_test[x2name]) + (b3 * XSTD_test[x3name])], axis=1) yXypred_test = yXypred_test.rename(columns={0: 'ypred'}) yXypred_test = pd.concat([yXypred_test, yXypred_test[yname] - yXypred_test['ypred']], axis=1) yXypred_test = yXypred_test.rename(columns={0: 'residual'}) yXypred_test.to_csv('yXypred_test.csv', header=True, index=False) #print(yXypred_test.columns) #Index(['y', 'x1', 'x2', 'x3', 'ypred'], dtype='object') ########## plot: test data (x1, y) and predicted data (x1, ypred) plt.figure(figsize=(8, 8)) # plt.scatter(yXypred_test[x1name], yXypred_test[yname], color = 'blue', label='Test Data') # plt.scatter(yXypred_test[x1name], yXypred_test['ypred'], color = 'red', label='Predicted Data') # plt.legend(bbox_to_anchor=(1, 0), loc='lower right', borderaxespad=1, fontsize=10) plt.title(x1name + ": Test Data, " + yname + ": Test Data and Predicted Data (based on Training Data)") plt.xlabel(x1name) plt.ylabel(yname) plt.grid(True) # plt.text(min(yXypred_test[x1name]), max(yXypred_test[yname]) * 1.00, "y = b0 + (b1 * x1) + (b2 * x2) + (b3 * x3)", size = 10, color = "black") plt.text(min(yXypred_test[x1name]), max(yXypred_test[yname]) * 0.90, "b0 = " + str(b0), size = 10, color = "black") plt.text(min(yXypred_test[x1name]), max(yXypred_test[yname]) * 0.80, "b1 = " + str(b1), size = 10, color = "black") plt.text(min(yXypred_test[x1name]), max(yXypred_test[yname]) * 0.70, "b2 = " + str(b2), size = 10, color = "black") plt.text(min(yXypred_test[x1name]), max(yXypred_test[yname]) * 0.60, "b3 = " + str(b3), size = 10, color = "black") # plt.savefig('Fig_03_2a.png') plt.show() ########## plot: test data (x1) and residual data (y - ypred) plt.figure(figsize=(8, 8)) # plt.scatter(yXypred_test[x1name], yXypred_test['residual'], color = 'green', label='Residual (actual - predicted)') # plt.legend(bbox_to_anchor=(1, 0), loc='lower right', borderaxespad=1, fontsize=10) plt.title(x1name + ": Training Data, " + 'residual' + ": Residual (actual - predicted)") plt.xlabel(x1name) plt.ylabel('Residual') plt.grid(True) # plt.savefig('Fig_03_2b.png') plt.show() |
stpkg04.py
| #################### Multiple Linear Regression ########## Run this code as follows #python3 stpkg04.py yXSTD.csv y x1 3 #python3 stpkg04.py (data file to load) (dependent variable/target in yXSTD.csv) (one specified independent variables in yXSTD.csv for polynominal linear regression) ########## import import sys import pandas as pd from sklearn.model_selection import train_test_split from sklearn.linear_model import LinearRegression from sklearn.preprocessing import PolynomialFeatures from sklearn.metrics import r2_score import matplotlib.pyplot as plt #print(sys.argv[0]) #stpkg04.py # dfname = sys.argv[1] yname = sys.argv[2] x1name = sys.argv[3] x1dim = int(sys.argv[4]) #x2name = sys.argv[4] #x3name = sys.argv[5] ########## ########## loading data #yXSTD = pd.read_csv('yXSTD.csv', header = 0) yXSTD = pd.read_csv(dfname, header = 0) #print(yXSTD.describe()) ''' y x1 x2 x3 count 100.000000 1.000000e+02 1.000000e+02 1.000000e+02 mean 1.008893 -1.232348e-16 1.576517e-16 -2.364775e-16 std 2.384226 1.005038e+00 1.005038e+00 1.005038e+00 min -4.001685 -2.592364e+00 -2.228172e+00 -2.412135e+00 25% -0.432215 -6.981616e-01 -7.997189e-01 -6.266296e-01 50% 0.755357 3.401995e-02 -5.543630e-02 1.232380e-02 75% 2.321260 6.719727e-01 7.398196e-01 6.364589e-01 max 6.808487 2.192663e+00 2.224031e+00 2.300629e+00 ''' XSTD = yXSTD.drop([yname], axis=1) #print(XSTD) #print(type(XSTD)) #X.to_csv('XSTD.csv', header=True, index=False) x1 = XSTD[x1name] x1 = pd.DataFrame(x1) #print(x1) #print(type(x1)) #<class 'pandas.core.frame.DataFrame'> y = yXSTD[yname] y = pd.DataFrame(y) #print(y) #print(type(y)) #y.to_csv('y.csv', header=True, index=False) ########## ########## creating training and test data x1_train, x1_test, y_train, y_test = train_test_split(x1, y, test_size=0.2, random_state = 0) #print(XSTD_train.shape) #print(XSTD_test.shape) #print(y_train.shape) #print(y_test.shape) ''' (80, 3) (20, 3) (80, 1) (20, 1) ''' #lr = linear_model.LinearRegression().fit(XSTD_train, y_train) #print(f"R2 of Training Data: {lr.score(XSTD_train, y_train):.4}") #print(f"R2 of Test Data (based on a model by Training Data): {lr.score(XSTD_test, y_test):.4}") ########## Polynominal Linear Regression (for Training Data) clf = LinearRegression() clf.fit(x1_train, y_train) #dimension #print(x1dim) pf = PolynomialFeatures(degree = x1dim, include_bias = False) x_poly = pf.fit_transform(x1_train) poly_reg = LinearRegression() poly_reg_fit = poly_reg.fit(x_poly, y_train) #print(poly_reg_fit.coef_) #[[2.05425368 0.11273714 0.03563863]] #print(type(poly_reg_fit.coef_)) #<class 'numpy.ndarray'> #print(poly_reg_fit.coef_[0][0]) #2.0542536782376684 # #print(poly_reg_fit.intercept_) #[0.99017672] polypred = poly_reg.predict(x_poly) #predicted y #ypred = clf.predict(y_train) #ypred = clf.predict(x1_train) #print(ypred) #print(type(ypred)) #<class 'numpy.ndarray'> ##### output data ypred_train = poly_reg_fit.intercept_[0] + (poly_reg_fit.coef_[0][0] * x1_train) + (poly_reg_fit.coef_[0][1] * (x1_train ** 2)) + (poly_reg_fit.coef_[0][2] * (x1_train **3)) ypred_train = ypred_train.rename(columns={x1name: 'ypred'}) #print(ypred_test) yx1ypred_train = pd.concat([y_train, x1_train, ypred_train], axis=1) yx1ypred_train.to_csv('yx1ypred_train.csv', header=True, index=False) yx1ypredres_train = pd.concat([yx1ypred_train, yx1ypred_train[yname] - yx1ypred_train['ypred']], axis=1) yx1ypredres_train = yx1ypredres_train.rename(columns={0: 'residual'}) yx1ypredres_train.to_csv('yx1ypredres_train.csv', header=True, index=False) ##### plot plt.scatter(yx1ypredres_train[x1name], yx1ypredres_train[yname], c = 'blue', label = 'Training Data') plt.scatter(yx1ypredres_train[x1name], yx1ypredres_train['ypred'], c = 'red', label = 'Predicted Data') # plt.text(min(yx1ypredres_train[x1name]), max(yx1ypredres_train[yname]) * 1.00, "y = b0 + (b1 * x1) + (b2 * x1^2) + (b3 * x1^3)", size = 10, color = "black") plt.text(min(yx1ypredres_train[x1name]), max(yx1ypredres_train[yname]) * 0.90, "b0 = " + str(poly_reg_fit.intercept_[0]), size = 10, color = "black") plt.text(min(yx1ypredres_train[x1name]), max(yx1ypredres_train[yname]) * 0.80, "b1 = " + str(poly_reg_fit.coef_[0][0]), size = 10, color = "black") plt.text(min(yx1ypredres_train[x1name]), max(yx1ypredres_train[yname]) * 0.70, "b2 = " + str(poly_reg_fit.coef_[0][1]), size = 10, color = "black") plt.text(min(yx1ypredres_train[x1name]), max(yx1ypredres_train[yname]) * 0.60, "b3 = " + str(poly_reg_fit.coef_[0][2]), size = 10, color = "black") plt.text(min(yx1ypredres_train[x1name]), max(yx1ypredres_train[yname]) * 0.50, "R^2={}".format(r2_score(y_train, polypred)), size = 10, color = "black") # plt.xlabel(x1name + ': Training Data') plt.ylabel(yname) plt.grid(True) # #plt.legend() plt.legend(bbox_to_anchor=(1, 0), loc='lower right', borderaxespad=1, fontsize=10) plt.title("Polynominal Regression: " + x1name) plt.savefig('Fig_04_1a.png') plt.show() ''' #plt.scatter(x1_train, y_train, c = 'blue', label = "R^2={}".format(r2_score(y_train, polypred))) plt.scatter(x1_train, y_train, c = 'blue', label = 'Training Data') plt.scatter(x1_train, polypred, c = 'red', label = 'Predicted Data') #plt.plot(x1_train, polypred, c = 'red') # plt.text(min(x1_train[x1name]), max(y_train[yname]) * 1.00, "y = b0 + (b1 * x1) + (b2 * x1^2) + (b3 * x1^3)", size = 10, color = "black") plt.text(min(x1_train[x1name]), max(y_train[yname]) * 0.90, "b0 = " + str(poly_reg_fit.intercept_[0]), size = 10, color = "black") plt.text(min(x1_train[x1name]), max(y_train[yname]) * 0.80, "b1 = " + str(poly_reg_fit.coef_[0][0]), size = 10, color = "black") plt.text(min(x1_train[x1name]), max(y_train[yname]) * 0.70, "b2 = " + str(poly_reg_fit.coef_[0][1]), size = 10, color = "black") plt.text(min(x1_train[x1name]), max(y_train[yname]) * 0.60, "b3 = " + str(poly_reg_fit.coef_[0][2]), size = 10, color = "black") plt.text(min(x1_train[x1name]), max(y_train[yname]) * 0.50, "R^2={}".format(r2_score(y_train, polypred)), size = 10, color = "black") # plt.xlabel(x1name + ': Training Data') plt.ylabel(yname) plt.grid(True) # #plt.legend() plt.legend(bbox_to_anchor=(1, 0), loc='lower right', borderaxespad=1, fontsize=10) plt.title("Polynominal Regression: " + x1name) plt.savefig('Fig_04_1a.png') plt.show() ''' ##### residual plot plt.scatter(yx1ypredres_train[x1name], yx1ypredres_train['residual'], c = 'green', label = 'Redisual: Training Data') # plt.xlabel(x1name + ': Training Data') plt.ylabel(yname) plt.grid(True) # #plt.legend() plt.legend(bbox_to_anchor=(1, 0), loc='lower right', borderaxespad=1, fontsize=10) plt.title("Polynominal Regression, Residual: " + x1name) plt.savefig('Fig_04_1b.png') plt.show() ''' ypred = pd.DataFrame(ypred) #print(ypred) #print(type(ypred)) #<class 'pandas.core.frame.DataFrame'> ypred = ypred.rename(columns={0: 'ypred'}) # #print(ypred) # #print(y_train) #print(type(y_train)) # #print(x1_train) #print(type(x1_train)) y_train.reset_index(drop=True, inplace=True) y_train = y_train.rename(columns={0: 'y'}) #print(y_train) x1_train.reset_index(drop=True, inplace=True) x1_train = x1_train.rename(columns={0: 'x1'}) #print(x1_train) #yx1ypred_train = pd.concat([y_train, x1_train, ypred], axis=1, ignore_index = True) yx1ypred_train = pd.concat([y_train, x1_train, ypred], axis=1) yx1ypred_train.to_csv('yx1ypred_train.csv', header=True, index=False) yx1ypredres_train = pd.concat([yx1ypred_train, yx1ypred_train[yname] - yx1ypred_train['ypred']], axis=1) yx1ypredres_train = yx1ypredres_train.rename(columns={0: 'residual'}) yx1ypredres_train.to_csv('yx1ypredres_train.csv', header=True, index=False) plt.scatter(yx1ypredres_train[x1name], yx1ypredres_train['residual'], c = 'green', label = 'Redisual: Training Data') #plt.scatter(x1_train, polypred, c = 'red', label = 'Predicted Data') #plt.plot(x1_train, polypred, c = 'red') # #plt.text(min(x1_train[x1name]), max(y_train[yname]) * 1.00, "y = b0 + (b1 * x1) + (b2 * x1^2) + (b3 * x1^3)", size = 10, color = "black") #plt.text(min(x1_train[x1name]), max(y_train[yname]) * 0.90, "b0 = " + str(poly_reg_fit.intercept_[0]), size = 10, color = "black") #plt.text(min(x1_train[x1name]), max(y_train[yname]) * 0.80, "b1 = " + str(poly_reg_fit.coef_[0][0]), size = 10, color = "black") #plt.text(min(x1_train[x1name]), max(y_train[yname]) * 0.70, "b2 = " + str(poly_reg_fit.coef_[0][1]), size = 10, color = "black") #plt.text(min(x1_train[x1name]), max(y_train[yname]) * 0.60, "b3 = " + str(poly_reg_fit.coef_[0][2]), size = 10, color = "black") #plt.text(min(x1_train[x1name]), max(y_train[yname]) * 0.50, "R^2={}".format(r2_score(y_train, polypred)), size = 10, color = "black") # plt.xlabel(x1name + ': Training Data') plt.ylabel(yname) plt.grid(True) # #plt.legend() plt.legend(bbox_to_anchor=(1, 0), loc='lower right', borderaxespad=1, fontsize=10) plt.title("Polynominal Regression, Residual: " + x1name) plt.savefig('Fig_04_1b.png') plt.show() ''' ########## Polynominal Linear Regression (for Test Data) ##### output data #x1_test #y_test ypred_test = poly_reg_fit.intercept_[0] + (poly_reg_fit.coef_[0][0] * x1_test) + (poly_reg_fit.coef_[0][1] * (x1_test ** 2)) + (poly_reg_fit.coef_[0][2] * (x1_test **3)) ypred_test = ypred_test.rename(columns={x1name: 'ypred'}) #print(ypred_test) yx1ypred_test = pd.concat([y_test, x1_test, ypred_test], axis=1) yx1ypred_test.to_csv('yx1ypred_test.csv', header=True, index=False) yx1ypredres_test = pd.concat([yx1ypred_test, yx1ypred_test[yname] - yx1ypred_test['ypred']], axis=1) yx1ypredres_test = yx1ypredres_test.rename(columns={0: 'residual'}) yx1ypredres_test.to_csv('yx1ypredres_test.csv', header=True, index=False) ##### plot plt.scatter(yx1ypredres_test[x1name], yx1ypredres_test[yname], c = 'blue', label = 'Test Data') plt.scatter(yx1ypredres_test[x1name], yx1ypredres_test['ypred'], c = 'red', label = 'Predicted Data') # plt.text(min(yx1ypredres_test[x1name]), max(yx1ypredres_test[yname]) * 1.00, "y = b0 + (b1 * x1) + (b2 * x1^2) + (b3 * x1^3)", size = 10, color = "black") plt.text(min(yx1ypredres_test[x1name]), max(yx1ypredres_test[yname]) * 0.90, "b0 = " + str(poly_reg_fit.intercept_[0]), size = 10, color = "black") plt.text(min(yx1ypredres_test[x1name]), max(yx1ypredres_test[yname]) * 0.80, "b1 = " + str(poly_reg_fit.coef_[0][0]), size = 10, color = "black") plt.text(min(yx1ypredres_test[x1name]), max(yx1ypredres_test[yname]) * 0.70, "b2 = " + str(poly_reg_fit.coef_[0][1]), size = 10, color = "black") plt.text(min(yx1ypredres_test[x1name]), max(yx1ypredres_test[yname]) * 0.60, "b3 = " + str(poly_reg_fit.coef_[0][2]), size = 10, color = "black") #plt.text(min(yx1ypredres_test[x1name]), max(yx1ypredres_test[yname]) * 0.50, "R^2={}".format(r2_score(y_train, polypred)), size = 10, color = "black") # plt.xlabel(x1name + ': Test Data') plt.ylabel(yname) plt.grid(True) # #plt.legend() plt.legend(bbox_to_anchor=(1, 0), loc='lower right', borderaxespad=1, fontsize=10) plt.title("Polynominal Regression: " + x1name) plt.savefig('Fig_04_2a.png') plt.show() ##### residual plot plt.scatter(yx1ypredres_test[x1name], yx1ypredres_test['residual'], c = 'green', label = 'Redisual: Test Data') # plt.xlabel(x1name + ': Test Data') plt.ylabel(yname) plt.grid(True) # #plt.legend() plt.legend(bbox_to_anchor=(1, 0), loc='lower right', borderaxespad=1, fontsize=10) plt.title("Polynominal Regression, Residual: " + x1name) plt.savefig('Fig_04_2b.png') plt.show() |
Figures










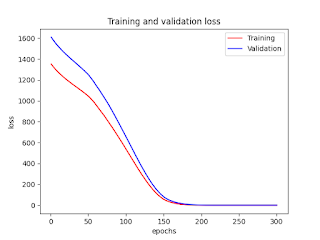

No comments:
Post a Comment